By Casey Mallinckrodt, Andrew Mellon Assistant Conservator
Conservators carry out technical analyses to identify materials to guide our decision-making, if treatment is required, and to predict and prevent future deterioration by ensuring that objects are handled and stored appropriately. In addition, technical information contributes to our understanding of an object within its cultural context and enhances what we can share with the museum public. This aspect is particularly important in the stewardship of historic arts of Africa because the information allows us, in partnership with our curatorial colleagues, to repopulate the histories of these objects with cultural information that was often lost when they were collected.
Historic arts of Africa often incorporate leather, fur, feathers, horn, and other proteinaceous materials, and ritual objects may be coated or anointed with mixtures that may contain blood, spit, or feces. Through the Andrew W. Mellon Foundation-funded Conservation Initiative for African Art, we have been able to collaborate with several scientists including Dr. Kristina Nelson, Director of the Chemical and Proteomic Mass Spectrometry Core Facility at Virginia Commonwealth University, to identify animal species from minute samples of leather, horn, feathers, and bone, and to determine whether blood or other proteinaceous materials are present in mixtures applied as coatings or as ritual libations.
Spectroscopic analysis and identification of proteins has been developed for multiple medical and life sciences applications. Diagnosis and screening for inherited metabolic diseases and identifying the source microbes in fungal and bacterial infections are just two examples. Two processes that have been used within art conservation are Liquid Chromatography-Tandem Mass Spectroscopy (LC-MS/MS) peptide sequencing, which Dr. Nelson used for our materials, and Matrix-Assisted Laser Desorption/Ionization-Time of Flight (MALDI-TOF) protein mass fingerprinting.

VMFA conservators Kate Gabrielli and Ainslie Harrison, and Dr.Kristina Nelson, Director, VCU Mass Spectrometry Facility
In the LC-MS/MS process that Dr. Nelson used to analyze our samples, the sample is enzymatically digested and peptides are separated from the other components of the sample using Liquid Chromatography- the “LC” of LC-MS/MS. As peptides elute off the LC column, their mass is measured. Additionally, each peptide is fragmented into a predictable chain of amino acids. An extensive online protein database, in this case the Uniprot database, is used to compare the amino acids to identify matches with known examples. The peptide and protein matches can be viewed in Scaffold software.
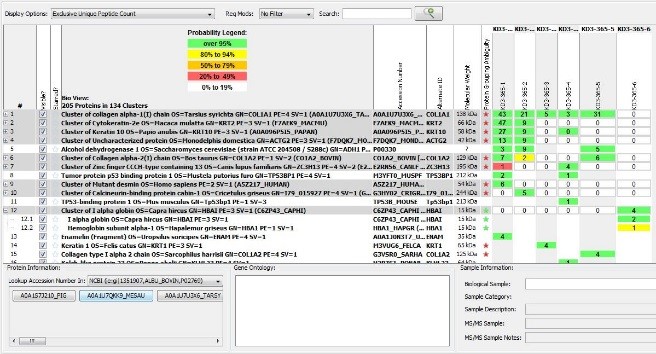
Screenshot of analytic results viewed in SCAFFOLD software.
The LC-MS/MS method is particularly well suited to identifying proteins within complex mixtures that are typical of African objects like the Adja Bocio (VMFA 2002.526a-e) with layers of materials encrusted over many years of ritual application.


Adja, Bocio, 19th-20th C. Benin, VMFA 2002.526 a-e overall (l) and surface detail of one side of the Janus head (r)
Although this dark crust on the head is primarily a plant resin with minerals and plant fibers mixed into it, peptide sequencing identified goat blood, consistent with the Vodun practice of animal sacrifice. Without this technology it would be difficult to identify blood and not possible for us to determine the source species.
The results can be precise and provide confirmation of a known practice or add new and important information to our understanding of an object and its history. But accurate interpretation of the results requires the specialized expertise of a scientist in partnership with the conservator’s and curator’s knowledge of the object and cultural context.
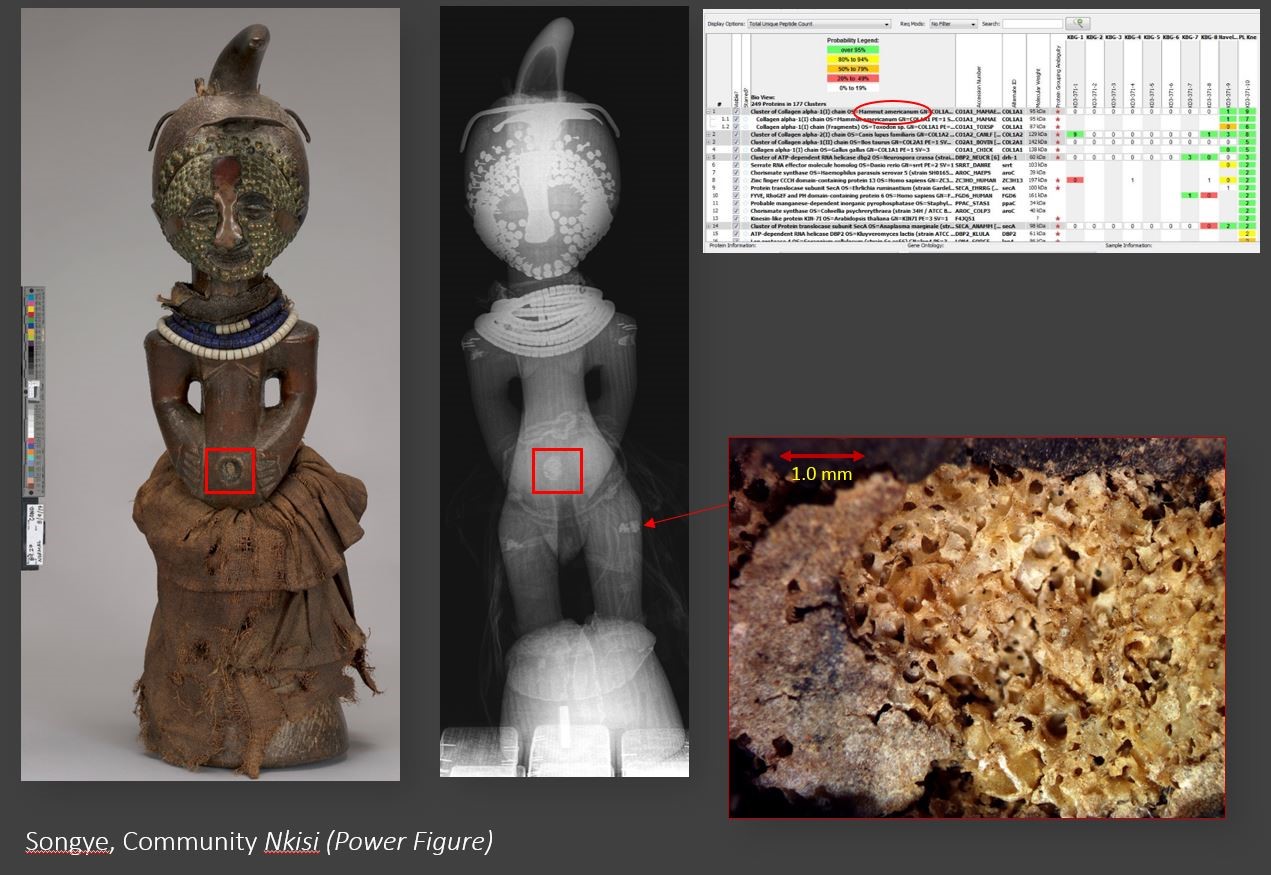
Peptide analysis of a material imbedded into the abdomen and sides of this Songye Community Nkisi (VMFA 89.27)
Peptide analysis of a material imbedded into the abdomen and sides of this Songye Community Nkisi (VMFA 89.27) identified it as bone; an exciting and rich addition to our understanding of the object. But the best species match was for American Mastodon – an animal confined to North America before becoming extinct approximately 10,500 years ago.

American Mastodon
While this might be interpreted as a major new finding, our collective knowledge suggests that the more prosaic and less robust matches for cow or dog are far more likely. Although the protein database is vast, results may be weighted toward species that are more extensively studied or to illegally trafficked animals. And animals from Central Africa may be less well documented, causing misidentifications.
Mastodons aside, this is a powerful and refined technology and the opportunity to work with Dr. Nelson has increased our knowledge of the objects we are studying and the information we are able to contribute to the literature on African collections.
Conservation projects utilizing peptide analysis.